BACK TO RESEARCH WITH IMPACT: FNR HIGHLIGHTS
AI developments have led to improvements in disease research by enabling new approaches in the study of genes, molecules and proteins – scientists are now tackling the challenge of building reliable AI tools.

It is well established that several diseases, cancers and genetic disorders, are linked to changes in genes, molecules, or proteins. Still, the exact connection is still to be found, as well as how to predict the behaviour of these elements, slowing down drug development and limiting treatment options for millions of people.
AI has significantly improved how genes, molecules, and proteins are studied: Thanks to AI, it has become easier to predict protein structures, suggest new drug candidates, as well as help design molecules with specific properties.
“Graph-based models have made it easier to understand complex biological data by representing it in structured ways. These tools have helped scientists gain deeper insights, test ideas faster, and reduce the need for expensive experiments. As a result, research and development in areas like drug discovery, genetic analysis, and disease understanding has become more efficient and precise. ”Zhiqiang Zhong Postdoc and computer scientist at the University of Luxembourg.
Challenge: Making sense of datasets
A key challenge, however: making sense of complex biological data that is often incomplete. Genes, molecules, and proteins all behave differently across conditions – a struggle for AI models. Therefore, building tools that are both accurate and reliable in real-world settings remains a major scientific goal.
“Many biological datasets are small, noisy, or not well-labelled, which makes it hard for AI models to learn useful patterns. At the same time, the systems we study—like genes, molecules, and proteins—are highly complex. These issues limit the accuracy and trustworthiness of current tools used in science and medicine.”
Tools to improve study of biological systems
Zhiqiang Zhong’s research uses AI to support researchers in predicting gene activity, as well as designing new molecules, and understanding protein properties. The tools being developed help improve the study of biological systems, ultimately supporting faster and more targeted drug development – reducing cost and uncertainty in biomedical research.
“I develop AI models that use existing biological knowledge in various forms—such as unstructured texts, structured databases, or semi-structured sources—to improve learning from limited or noisy data.”
“My work focuses on building graph-based models that capture the structure of molecules, proteins, and gene-related data. These models help computers make better predictions and reduce the need for large, high-quality datasets. I also work on making these tools easier to interpret, so that scientists can trust the results. This enables faster and more reliable exploration of new questions in biology and medicine. ”Zhiqiang Zhong Postdoc and computer scientist at the University of Luxembourg.
Better predictions thanks to combination of biological data and large language models
Zhiqiang Zhong’s research has shown that combining large language models with biological data makes for better predictions in molecular and protein-related tasks. The computer scientist has also developed graph-based and interpretable AI methods that assist scientists in understanding model outputs, making these tools more usable and trustworthy in real-world biomedical research.
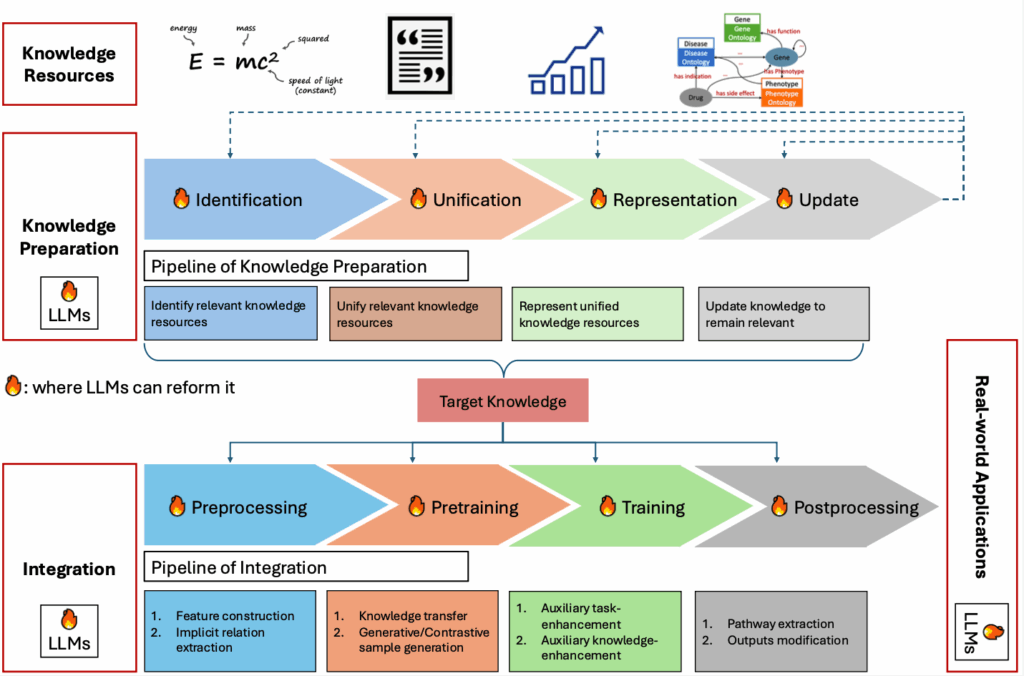

Dr Zhiqiang Zhong is a Marie Skłodowska-Curie Actions (MSCA) Postdoctoral Researcher, YIA (Young International Academics) Fellow and computer scientist at the University of Luxembourg. His PhD was supported by one of the first FNR PRIDE grants (DTU: ‘Security and Privacy for System Protection’)
MORE ABOUT ZHIQIANG ZHONG
Describing his research in one sentence
“I build AI tools that help scientists better understand genes, molecules, and proteins, so they can develop new treatments and explore biological questions more effectively.”
Why he chose Luxembourg for his research
“I chose to do my research in Luxembourg because of its strong support for interdisciplinary and international collaboration. The country invests strategically in innovation, especially in data science, health, and digital technologies. The University of Luxembourg provides a dynamic research environment with close ties to public institutions and industry. I value the openness to new ideas and the focus on applied research that addresses real-world needs. Luxembourg’s small size also makes it easier to build strong cross-sector networks, which helps accelerate innovation and turn scientific progress into practical outcomes.”
Mentors with an impact
“My advisors Jun Pang and Davide Mottin have greatly influenced my research path. They encouraged independent thinking, supported interdisciplinary exploration, and emphasized the balance between scientific depth and real-world relevance—guidance that continues to shape how I approach research today.”
Where he sees himself in 5 years
“In five years, I see myself leading interdisciplinary research at the intersection of AI and biology—either in academia or a research-driven industry lab. I aim to develop AI tools that support scientific discovery, mentor young researchers, and connect academic innovation with real-world applications.”
Describing what kind of researcher he is
“I am a curious and solution-oriented researcher, motivated by the potential of AI to speed up scientific discovery. I chose this field to connect machine learning with biology, and joined the University of Luxembourg for its interdisciplinary research culture and strong support for collaborative, high-impact science.”
Why he loves science
“I love how science turns questions into knowledge and ideas into practical tools. Research gives me the freedom to explore, learn, and create solutions that can make a real difference. It is a continuous process of discovery that connects curiosity with real-world impact.”
On his research, peer to peer
“My research focuses on developing knowledge-augmented machine learning models for biological tasks. I integrate structured and unstructured biological knowledge into deep learning architectures to improve generalization in the presence of data sparsity. My recent work explores foundation models and contrastive learning for gene expression prediction, protein property analysis, and molecule generation. I also design post-hoc explanation methods to improve model interpretability. These approaches aim to create more robust and transparent AI systems for applications such as drug discovery, cell reprogramming, and biological network inference.”
Related highlights
Spotlight on Young Researchers: Ensuring sustainable water use for Luxembourg
In Luxembourg, nearly one tenth of water consumption happens in agriculture. Changing rainfall patterns and rising irrigation needs during summer…
Read more
Spotlight on Young Researchers – Revisited: From researcher to project manager
When Xianqing Mao was featured in Spotlight on Young Researchers in 2017, she had completed her medical degree and was…
Read more
Spotlight on Young Researchers: Giving robots a more human touch
The use of robots is rising, both in industry and in homes. This is made possible by scientists leveraging artificial…
Read more
Spotlight on Young Researchers: Cancer in older people & the need for a tailored approach
Cancer in adults aged 65+ is increasing, raising pressure on healthcare systems worldwide, creating economic and social burdens for families…
Read more
Spotlight on Young Researchers: A behavioural science approach to financial misconduct
The rapidly evolving risk landscape financial institutions navigate has brought uncertainty and a risk, making them vulnerable to misconduct and…
Read more
Spotlight on Young Researchers: An eco-friendlier approach to plastics
Polyurethanes (PUs) are widely used plastics, produced in large quantities each year, and found in products like foams, coatings, sporting…
Read more